Story by: Melinda Ballengee —
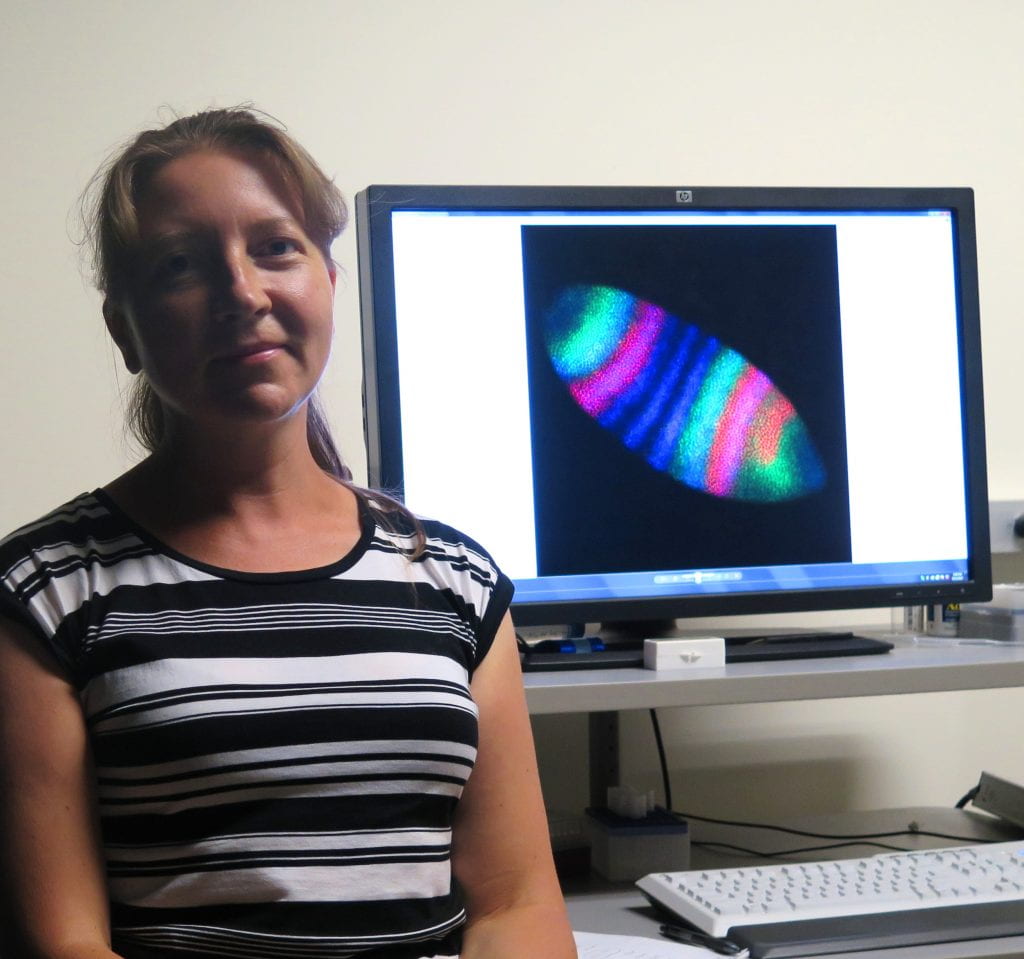
Meet Svetlana Surkova, a J-1 research fellow who has come to USC to study how gene expression determines spatial organization in early development organisms. She comes to us by way of St. Petersburg Polytechnical University (SPbSPU) in Russia. Her laboratory has established a long-term collaboration with USC’s group lead by Sergey Nuzhdin in the program of Molecular and Computational Biology. Sveta is the 4th SPbSPU visitor to Sergey’s lab; likewise USC’s Stephan Haas, Tanya Tatarinova, and Josh Shiffman visited SPbSPU to deliver lectures and engage in reserach collaborations. These ‘mirror’ activities are supported by a combination of NIH UO1: ‘Multiscale Modelling’ grant to Sergey Nuzhdin, Paul Marjoram and Remo Rohs in USC; as well as 5/100 and Russian National Science Foundation grants to Sergey Nuzhdin and Maria Samsonova.
Svetlana, in her second trip to USC, uses fruit flies as a model organism/network of genes to conduct analysis of spatial and temporal precision of gene expression. Through the use of imagining techniques she is able to study gene expression at the level of individual molecules. I asked Svetlana what attracted her to this line of research. “I like this project because it gives a possibility to combine novel experimental techniques, high- resolution imaging and 3D data processing. It enables me to apply different skills and to learn new methods.”
I was curious to know how humans can benefit by her research. “Genes involved in human disease can be identified using genome-wide association studies. This method searches the genome for small variations that occur more frequently in people with a particular disease than in people without the disease. However, it was shown that the revealed genetic variation cannot fully explain all variation in phenotypes. In our research we use different fruit fly genotypes to study how the differences in sequencing affect gene expression patterns or ‘molecular phenotypes’.” NIH recognizes this research as a new paradigm for building genotype-to-phenotype map – a concept on which precision medicine of the future will be built. Using concepts developed in this work, we will learn how abnormalities of development originate from stochastic nature of gene interactions – the conceptual knowledge required for better treatments.